Backes' Laboratory
Research Interests
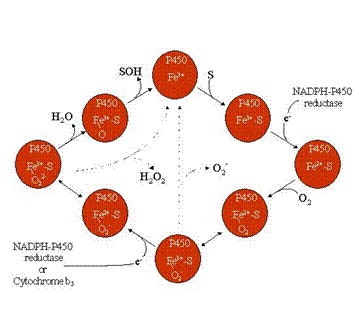
Cytochrome P450 refers to a superfamily of heme-containing enzymes involved in the metabolism of a wide variety of drugs, hydrocarbons, and other foreign compounds. This enzyme system is capable of metabolizing substrates of widely varying structure, with the basic process to insert an oxygen atom into the substrate molecule. The resulting product is more water soluble and generally more rapidly excreted; however, some of the resulting metabolites are reactive and can covalently bind to DNA, RNA and proteins. The bioactivation of foreign compounds by cytochrome P450 and some other xenobiotic-metabolizing enzymes can lead to toxicity, mutagenesis, carcinogenesis, and birth defects.
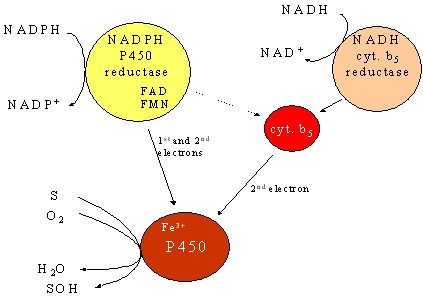
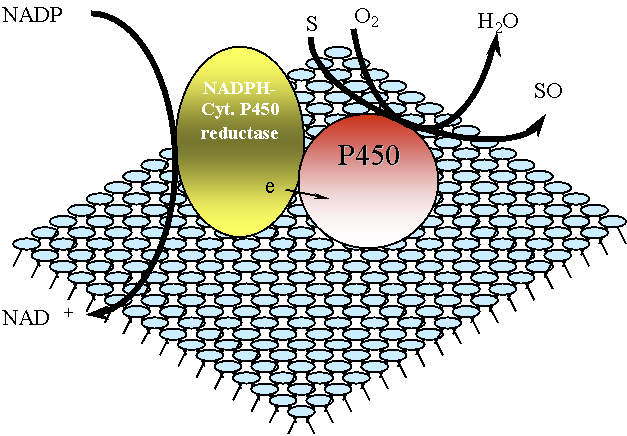
Figure 1 - Schematic of the cytochrome P450 enzyme system, showing the substrates and necessary co-factors in addition to the NADPH-cytochrome P450 reductase - P450 interaction that represents the simplest catalytically competent NADPH-mediated system
There are two reasons the cytochrome P450 system can metabolize a wide variety of substrates. First, the active site of cytochrome P450 is relatively non-selective and is capable of accommodating substrates of diverse chemical structures. The second reason for its broad selectivity is that there are many different P450 enzymes, each with their own characteristic substrate selectivities. Each of the P450 enzymes is independently regulated at least in part, by nutritional status, and prior exposure to foreign compounds.
The enzymes of the cytochrome P450 system do not act alone, but are part of an electron transport chain found in the endoplasmic reticulum. P450 requires an interaction with NADPH-cytochrome P450 reductase, a flavoprotein that is necessary for the transfer the reducing equivalents to P450. Other proteins that interact with those of the P450 system include cytochrome b5, NADH-cytochrome b5 reductase, and heme oxygenase.
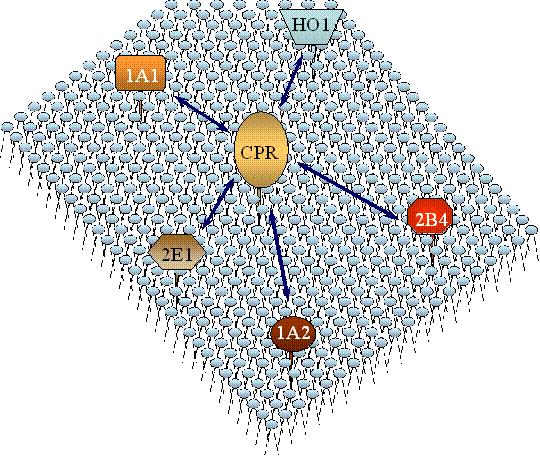
Figure 2 - Schematic of the competition among P450 enzymes for limiting reductase
found in the microsomal membrane. In liver microsomes the level of P450 exceeds that
of the reductase by a factor of 10:1 to 20:1. When considering the fact that there
are many different P450 enzymes present, there is the possibility for many different
possible types of interactions.
An important characteristic of the P450 system that can influence its catalytic effectiveness is that, in most tissues, P450 is present in a large molar excess over the reductase. This creates conditions where a particular P450 enzyme will need to effectively compete with other P450 enzymes for the available reductase or be metabolically silent. The resulting metabolism of a foreign compound by the P450 system will depend on which P450 enzymes most effectively receive electrons from the reductase. A diagram of the problem is shown where multiple P450 enzymes and other enzymes must compete for the reductase.
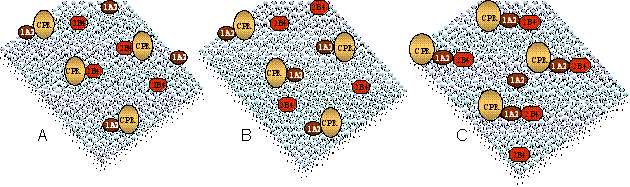
Figure 3 - Different mechanisms for the interaction of two P450 enzymes with reductase. When reductase concentrations are limiting, the ability of an individual P450 to compete for reductase will influence its catalytic effectiveness. Panel A shows a random distribution pattern for the reductase between two P450 isozymes. Under these conditions, both isozymes would be equally effective at competing for the reductase. Panel B shows the segregation of reductase to one of the P450 isozymes. Under these conditions, one enzyme (in this case 1A2) more effectively competes for the reductase. Panel C shows the formation of 1A2-2B4 complex that has altered binding characteristics when compared to the simple binary complexes. According to this model, the 1A2-2B4 complex preferentially binds reductase on the 1A2 moiety. This interaction differs from the type expected in the simple binary system (containing only reductase and a single P450), and can significantly influence the activity of this enzyme.